Categorization functions¶
Submodule pyiomica.categorizationFunctions
Example of use of a function described in this module:
# import the package and its mudule Categorization functions
import pyiomica as pio
from pyiomica import categorizationFunctions as cf
# Location of this example data
dir = pio.ConstantPyIOmicaExamplesDirectory
# Unzip sample data
with pio.zipfile.ZipFile(pio.os.path.join(dir, 'SLV.zip'), "r") as zipFile:
zipFile.extractall(path=dir)
# Process sample dataset SLV Hourly 1
dataName = 'SLV_Hourly1TimeSeries'
saveDir = pio.os.path.join('results', dataName, '')
dataDir = pio.os.path.join(dir, 'SLV')
df_data = pio.pd.read_csv(pio.os.path.join(dataDir, dataName + '.csv'), index_col=[0,1,2], header=0)
cf.calculateTimeSeriesCategorization(df_data, dataName, saveDir, NumberOfRandomSamples = 10**4)
cf.clusterTimeSeriesCategorization(dataName, saveDir)
cf.visualizeTimeSeriesCategorization(dataName, saveDir)
One of the figures generated by visualizeTimeSeriesCategorization is shown below:
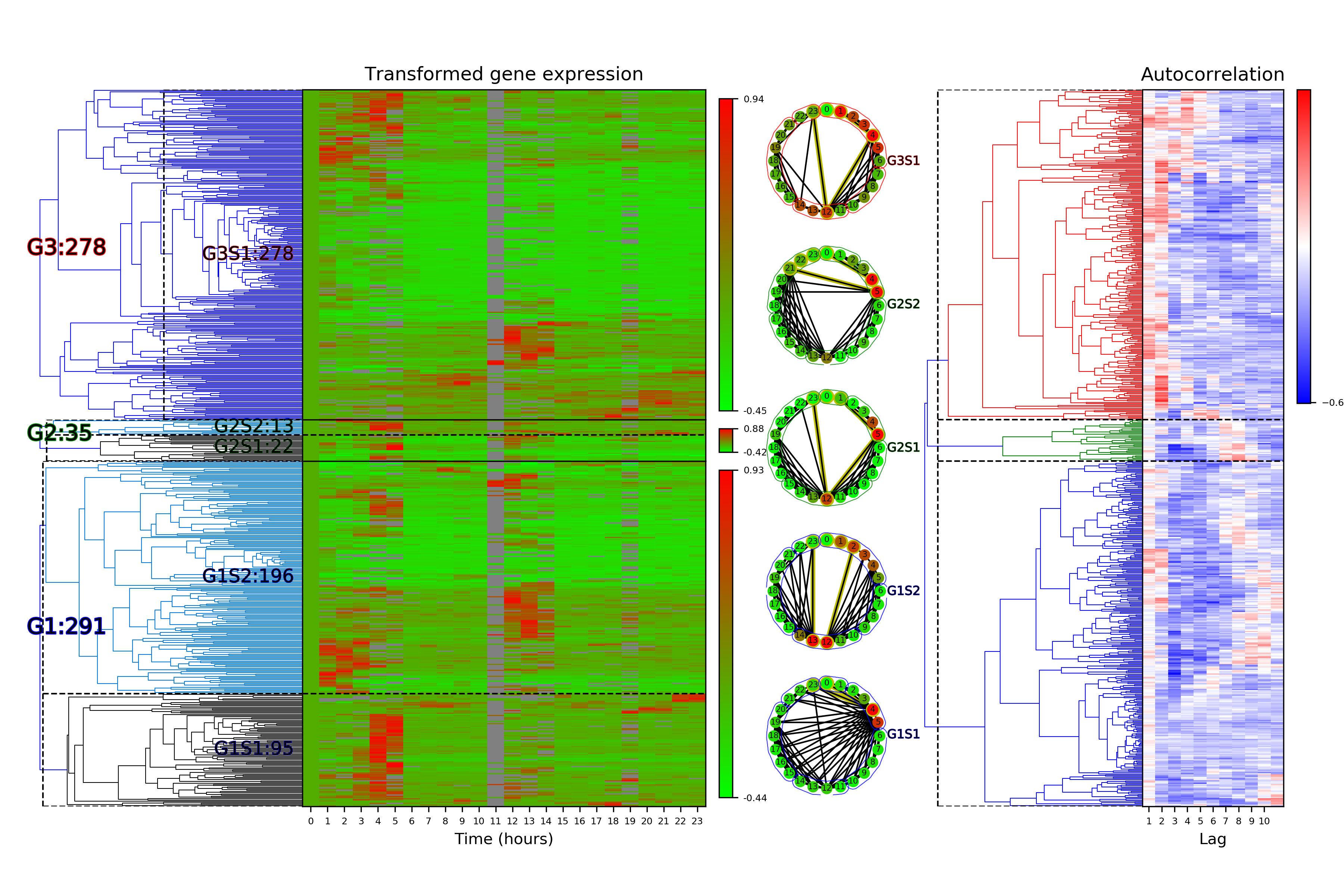
Categorization functions
Functions:
|
Time series classification. |
|
Visualize time series classification. |
|
Visualize time series classification. |
- calculateTimeSeriesCategorization(df_data, dataName, saveDir, hdf5fileName=None, p_cutoff=0.05, fraction=0.75, constantSignalsCutoff=0.0, lowValuesToTag=1.0, lowValuesToTagWith=1.0, NumberOfRandomSamples=100000, NumberOfCPUs=4, referencePoint=0, autocorrelationBased=True, calculateAutocorrelations=False, calculatePeriodograms=False, preProcessData=True)[source]¶
Time series classification.
- Parameters:
- df_data: pandas.DataFrame
Data to process
- dataName: str
Data name, e.g. “myData_1”
- saveDir: str
Path of directories poining to data storage
- hdf5fileName: str, Default None
Preferred hdf5 file name and location
- p_cutoff: float, Default 0.05
Significance cutoff signals selection
- fraction: float, Default 0.75
Fraction of non-zero point in a signal
- constantSignalsCutoff: float, Default 0.
Parameter to consider a signal constant
- lowValuesToTag: float, Default 1.
Values below this are considered low
- lowValuesToTagWith: float, Default 1.
Low values to tag with
- NumberOfRandomSamples: int, Default 10**5
Size of the bootstrap distribution to generate
- NumberOfCPUs: int, Default 4
Number of processes allowed to use in calculations
- referencePoint: int, Default 0
Reference point
- autocorrelationBased: boolean, Default True
Whether Autocorrelation of Frequency based
- calculateAutocorrelations: boolean, Default False
Whether to recalculate Autocorrelations
- calculatePeriodograms: boolean, Default False
Whether to recalculate Periodograms
- preProcessData: boolean, Default True
Whether to preprocess data, i.e. filter, normalize etc.
- Returns:
None
- Usage:
calculateTimeSeriesCategorization(df_data, dataName, saveDir)
- clusterTimeSeriesCategorization(dataName, saveDir, numberOfLagsToDraw=3, hdf5fileName=None, exportClusteringObjects=False, writeClusteringObjectToBinaries=True, autocorrelationBased=True, method='weighted', metric='correlation', significance='Elbow')[source]¶
Visualize time series classification.
- Parameters:
- dataName: str
Data name, e.g. “myData_1”
- saveDir: str
Path of directories pointing to data storage
- numberOfLagsToDraw: int, Default 3
First top-N lags (or frequencies) to draw
- hdf5fileName: str, Default None
HDF5 storage path and name
- exportClusteringObjects: boolean, Default False
Whether to export clustering objects to xlsx files
- writeClusteringObjectToBinaries: boolean, Default True
Whether to export clustering objects to binary (pickle) files
- autocorrelationBased: boolean, Default True
Whether to label to print on the plots
- method: str, Default ‘weighted’
Linkage calculation method
- metric: str, Default ‘correlation’
Distance measure
- significance: str, Default ‘Elbow’
Method for determining optimal number of groups and subgroups
- Returns:
None
- Usage:
clusterTimeSeriesClassification(‘myData_1’, ‘/dir1/dir2/’)
- visualizeTimeSeriesCategorization(dataName, saveDir, numberOfLagsToDraw=3, autocorrelationBased=True, xLabel='Time', plotLabel='Transformed Expression', horizontal=False, minNumberOfCommunities=2, communitiesMethod='WDPVG', direction='left', weight='distance')[source]¶
Visualize time series classification.
- Parameters:
- dataName: str
Data name, e.g. “myData_1”
- saveDir: str
Path of directories pointing to data storage
- numberOfLagsToDraw: boolean, Default 3
First top-N lags (or frequencies) to draw
- autocorrelationBased: boolean, Default True
Whether autocorrelation or frequency based
- xLabel: str, Default ‘Time’
X-axis label
- plotLabel: str, Default ‘Transformed Expression’
Label for the heatmap plot
- horizontal: boolean, Default False
Whether to use horizontal or natural visibility graph.
- minNumberOfCommunities: int, Default 2
Number of communities to find depends on the number of splits. This parameter is ignored in methods that automatically estimate optimal number of communities.
- communitiesMethod: str, Default ‘WDPVG’
- String defining the method to use for communitiy detection:
‘Girvan_Newman’: edge betweenness centrality based approach
‘betweenness_centrality’: reflected graph node betweenness centrality based approach
‘WDPVG’: weighted dual perspective visibility graph method (note to also set weight variable)
- direction:str, default ‘left’
- The direction that nodes aggregate to communities:
None: no specific direction, e.g. both sides.
‘left’: nodes can only aggregate to the left side hubs, e.g. early hubs
‘right’: nodes can only aggregate to the right side hubs, e.g. later hubs
- weight: str, Default ‘distance’
- Type of weight for communitiesMethod=’WDPVG’:
None: no weighted
‘time’: weight = abs(times[i] - times[j])
‘tan’: weight = abs((data[i] - data[j])/(times[i] - times[j])) + 10**(-8)
‘distance’: weight = A[i, j] = A[j, i] = ((data[i] - data[j])**2 + (times[i] - times[j])**2)**0.5
- Returns:
None
- Usage:
visualizeTimeSeriesClassification(‘myData_1’, ‘/dir1/dir2/’)